Epigenetic mechanisms controlling liver development and hepatic metabolic pathways
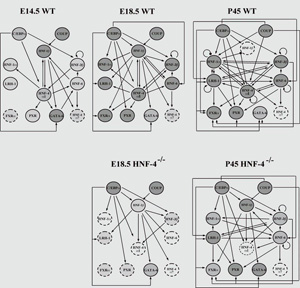 A network of tissue-restricted transcription factors specifies the genetic program that regulates endoderm differentiation and morphogenesis. Our previous studies have established that transcription factors LRH-1, HNF-3b, HNF-4, HNF-1b, HNF-6 and HNF-1a, form a transcriptional network via cross-regulatory and autoregulatory loops, which is crucial for the specification and maintenance of the hepatic phenotype. Research aimed at the understanding the mechanisms of how this small set of transcription factors play role in the developmental decisions of progenitor cells to differentiate into specific cell types, promises important new insights into the basic principles employed by living organisms that control cellular diversification. Our focus is on studying the mechanistic aspects of transcriptional activation with major emphasis placed on getting insights into the interplay between the recruitment of the pre-initiation complex components and changes in chromatin structure during liver development and upon exposure to various metabolic signals. We are studying the genes involved in the regulation of hepatic cholesterol-bile acid metabolism, fatty acid metabolism and glucose homeostasis using transgenic and knock-out animal models. Specific emphasis is given to mouse models lacking or overexpressing specific histone modifying enzymes. Many of our previous studies suggest that histone modifying enzymes exert their roles not only through modification of histones, but also through targeting transcription factors and other modification enzymes. These established an integrated concept of epigenetic regulatory pathways, whose roles in differentiation processes and cellular functions is being investigated.
A network of tissue-restricted transcription factors specifies the genetic program that regulates endoderm differentiation and morphogenesis. Our previous studies have established that transcription factors LRH-1, HNF-3b, HNF-4, HNF-1b, HNF-6 and HNF-1a, form a transcriptional network via cross-regulatory and autoregulatory loops, which is crucial for the specification and maintenance of the hepatic phenotype. Research aimed at the understanding the mechanisms of how this small set of transcription factors play role in the developmental decisions of progenitor cells to differentiate into specific cell types, promises important new insights into the basic principles employed by living organisms that control cellular diversification. Our focus is on studying the mechanistic aspects of transcriptional activation with major emphasis placed on getting insights into the interplay between the recruitment of the pre-initiation complex components and changes in chromatin structure during liver development and upon exposure to various metabolic signals. We are studying the genes involved in the regulation of hepatic cholesterol-bile acid metabolism, fatty acid metabolism and glucose homeostasis using transgenic and knock-out animal models. Specific emphasis is given to mouse models lacking or overexpressing specific histone modifying enzymes. Many of our previous studies suggest that histone modifying enzymes exert their roles not only through modification of histones, but also through targeting transcription factors and other modification enzymes. These established an integrated concept of epigenetic regulatory pathways, whose roles in differentiation processes and cellular functions is being investigated.
Studies on chromatin modifying and transcription complexes implicated in cancer pathogenesis
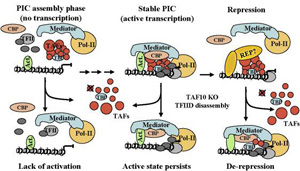 Carcinogenesis is a progression of events resulting from alterations in the processing of genetic information. These alterations result from stable genetic changes (mutations) involving tumor suppressor genes, or oncogenes and also from epigenetic changes, which are modifications in gene function without a change in DNA sequence. Epigenetic mechanisms often altered in cancer cells are histone modifications (acetylation, methylation, phosphorylation) and DNA methylation. These aberrant epigenetic mechanisms are manifested in both global changes in chromatin packaging and in localized changes in the binding of regulatory proteins to promoters that influence the transcription of genes important to the cancer process. Our studies are focused on histone methyltransferases, which have been implicated in the pathogenesis of hepatocellular carcinoma. We aim at the deeper understanding of the biological mechanisms responsible for cancer-specific expression patterns by the description of complete “regulomes” for the epigenetic modifiers.
Carcinogenesis is a progression of events resulting from alterations in the processing of genetic information. These alterations result from stable genetic changes (mutations) involving tumor suppressor genes, or oncogenes and also from epigenetic changes, which are modifications in gene function without a change in DNA sequence. Epigenetic mechanisms often altered in cancer cells are histone modifications (acetylation, methylation, phosphorylation) and DNA methylation. These aberrant epigenetic mechanisms are manifested in both global changes in chromatin packaging and in localized changes in the binding of regulatory proteins to promoters that influence the transcription of genes important to the cancer process. Our studies are focused on histone methyltransferases, which have been implicated in the pathogenesis of hepatocellular carcinoma. We aim at the deeper understanding of the biological mechanisms responsible for cancer-specific expression patterns by the description of complete “regulomes” for the epigenetic modifiers.
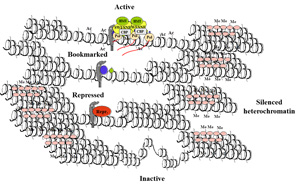 Our work combines global search data (factor occupancy, expression and chromatin modification maps) in a variety of relevant mouse models. A unique feature of our approach is that the studies involve analysis of the interplay of epigenetic modifiers with the general transcription machinery. The significance of the latter comes from the fact that most of the enzymes responsible for the epigenetic modifications play important functions in the transcriptional regulation of genes during normal mammalian development and during various signal transduction pathways.
Our work combines global search data (factor occupancy, expression and chromatin modification maps) in a variety of relevant mouse models. A unique feature of our approach is that the studies involve analysis of the interplay of epigenetic modifiers with the general transcription machinery. The significance of the latter comes from the fact that most of the enzymes responsible for the epigenetic modifications play important functions in the transcriptional regulation of genes during normal mammalian development and during various signal transduction pathways.
Importantly many of them are integral components of the transcription machinery required for proper temporal and spatial activation of genes in different cell types. Therefore we believe that studies towards the specific regulatory environment of proteins are essential to understand the complexity and disease-related alterations.




