A) Establishment of cellular fate
Neurogenesis in Drosophila takes place in two distinct developmental periods during embryonic and larval stages. Most neuronal cell lineages of the Central Nervous System (CNS) are produced via two asymmetric divisions. By the first division, each one of the Neuroblasts (NBs) [the Neuronal Stem Cells (NSCs) of Drosophila], generate a “renewed” NB and a short-lived precursor cell, the Ganglion Mother Cell (GMC). Upon the second division, the GMCs generate, primarily asymmetrically, two neurons of different fates or a neuron and a glia. In each of these two asymmetric divisions cell fate decisions are regulated by Delta-Notch dependent cell-cell signalling. In collaboration with Dr. Delidakis group, we have shown that the bHLH-O transcription factor HEY is a transcriptional target of Notch signalling and its effector molecule in GMC asymmetric cell divisions that establish an initial dichotomy in cellular fate within neuronal lineages (“A” type-Notch responsive- versus “B” type neurons). Ongoing research focuses on:
- The transcriptional and post-transcriptional regulation of Hey gene expression in Drosophila nervous system
- The identification of HEY transcriptional targets
- The elucidation of Hey expression and function in a non-neuronal Drosophila tissue, the developing and functional midgut
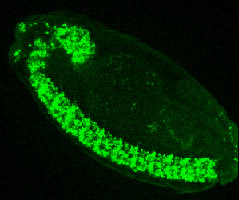
Expression of Hey gene in developing CNS and midgut primordia
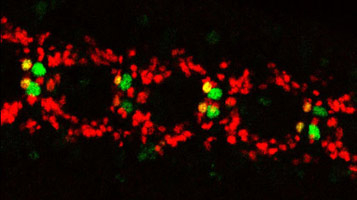
Expression of Hey (RED) in “A” type (vMP2) neurons of the MP2 neuronal lineage (GREEN)
B) Development of neurochemical specificity
The neurochemical specificity defines a terminal property of each neuron in a mature nervous system. This property relies on complex regulatory cascades and transcription factors (TFs) that establish the temporal and neuron-type specific expression of key neuropeptides and chemical transmitter biosynthetic enzymes that characterise terminally differentiated neurons. Our research centers on the regulation of Tyramine-β hydroxylase (Tβh) gene expression, which characterizes neuronal populations producing the neurotransmitter–neurohormone Octopamine. TBH is a key enzyme in the biosynthesis of Octopamine, and we developed tools to manipulate Octopamine metabolism in insects and study its effects on specific physiological processes, such as female reproductive activity and stress.
Using bioinformatic tools and immunocytochemical analysis, we now analyse cis regulatory regions of Tβh locus, investigating their impact on Tbh expression pattern in the embryonic ventral nerve cord and adult abdominal ganglia neurons that innervate female reproductive organs.
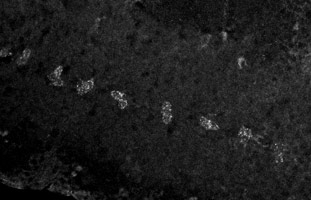
TBH-expressing neurons in embryonic CNS midline
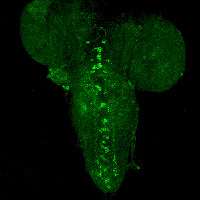
TBH-expressing neurons in larval CNS
C) Exploring Octopamine Neurotransmitter System in insects
The biogenic amine Octopamine is an insect neurotransmitter and neurohormone with functions similar to those of noradrenaline in vertebrates. Molecular genetic analysis of Drosophila genes involved in Octopamine biosynthesis has revealed its importance in various physiological processes, such as appetitive olfactory learning, sleep, female fertility, and stress reactivity. Our work has established the requirement of Octopamine for Drosophila female fertility, as Octopamine-deficient flies retain mature eggs and become sterile due to an ovulation defect. We now study the Octopamine system in insects of agricultural and medical importance (e.g., C. capitata and A. gambiae), using advanced methods to inactivate the Tβh gene. Our aim is to produce Octopamine-deficient insects to study reproductive effects and explore pest management applications.

Research on Octopamine-deficient insects




